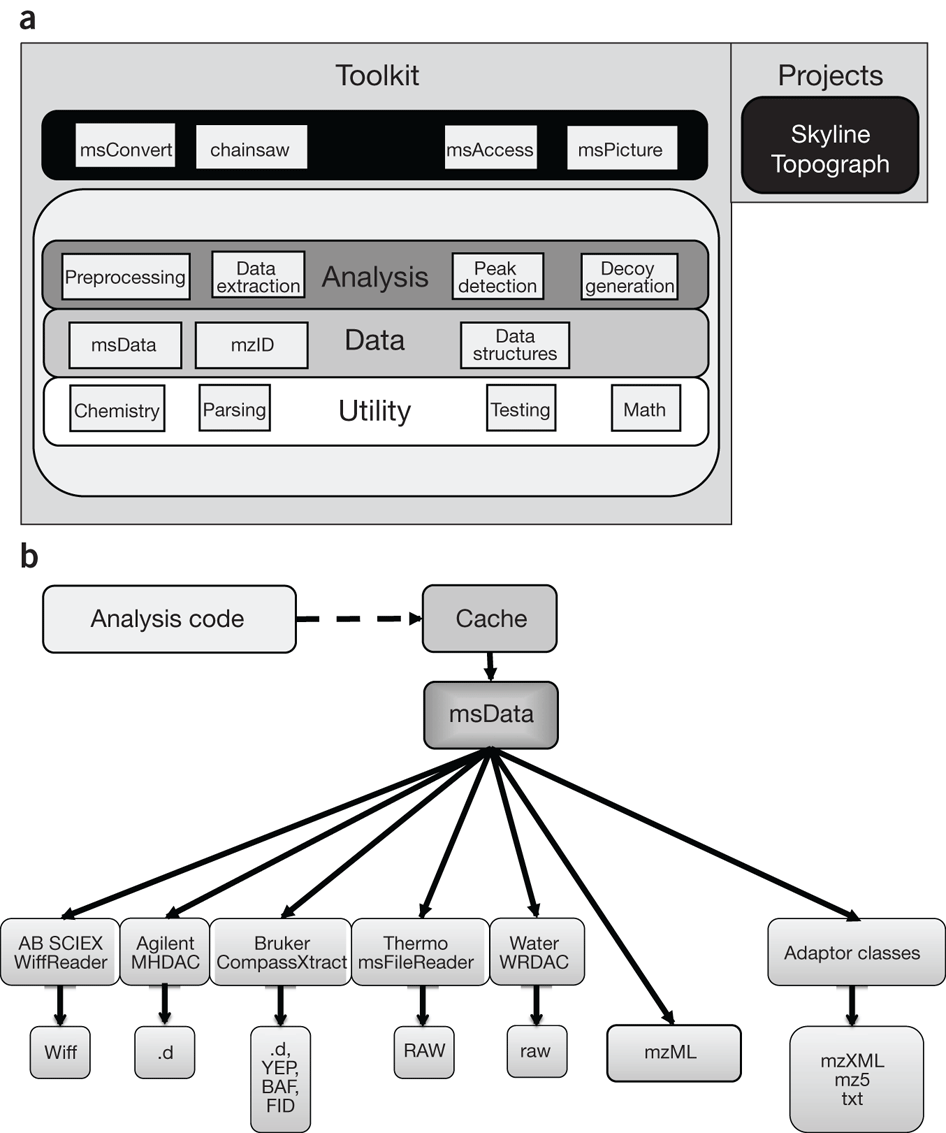
Cleavable Cross-Linkers and Mass Spectrometry for the Ultimate Task of Profiling Protein–Protein Interaction Networks in Vivo | Journal of Proteome Research
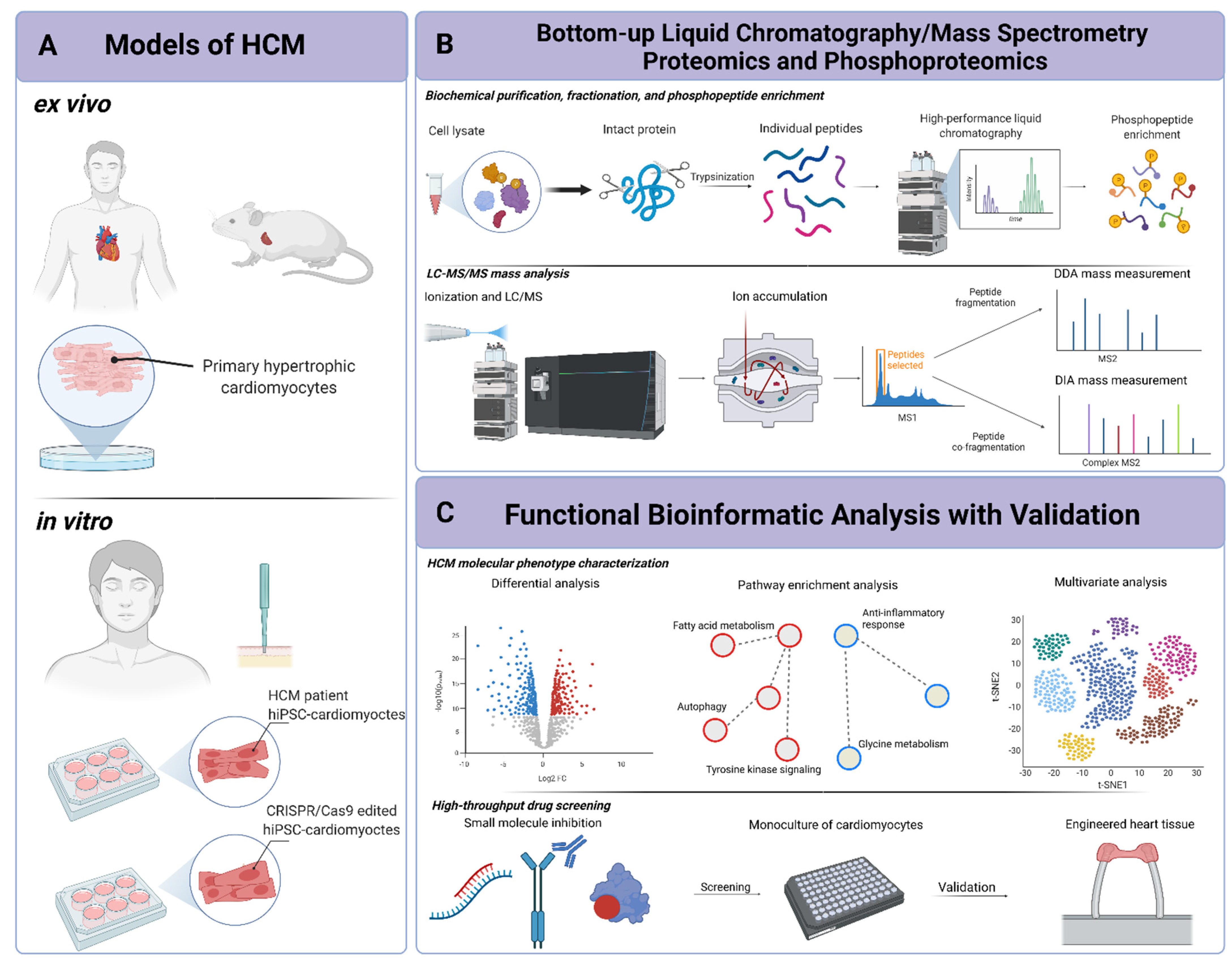
IJMS | Free Full-Text | Mass-Spectrometry-Based Functional Proteomic and Phosphoproteomic Technologies and Their Application for Analyzing Ex Vivo and In Vitro Models of Hypertrophic Cardiomyopathy
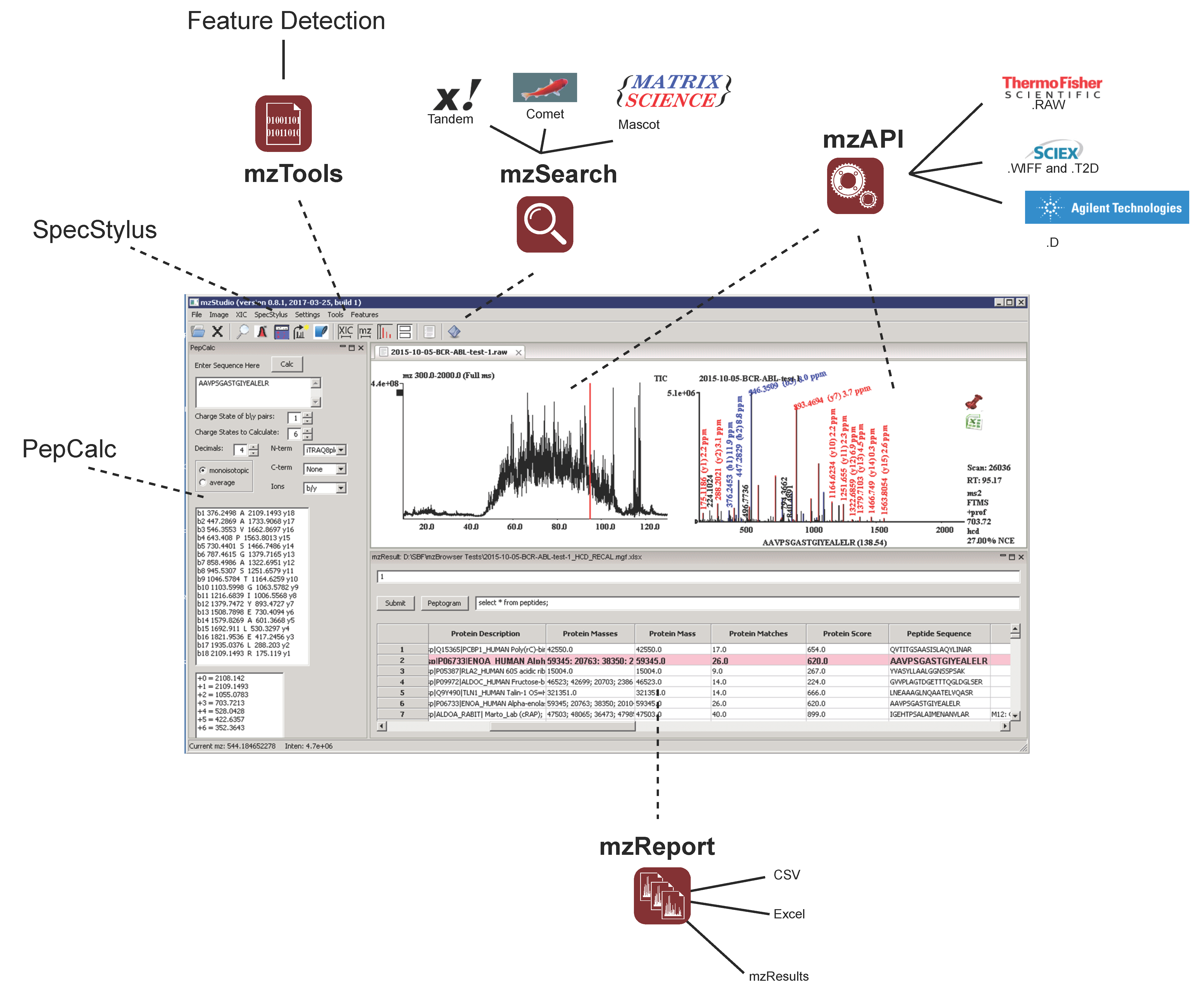
Proteomes | Free Full-Text | mzStudio: A Dynamic Digital Canvas for User-Driven Interrogation of Mass Spectrometry Data
![PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a99681b245131e0bc2523825fc0764e3de8c95c0/2-Table1-1.png)
PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar

Protein Dynamics in Solution by Quantitative Crosslinking/Mass Spectrometry: Trends in Biochemical Sciences
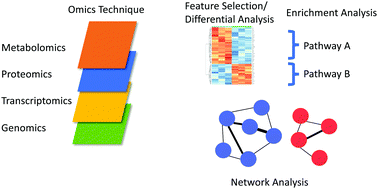
Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics–metabolomics analysis - Molecular Omics (RSC Publishing)

Native Mass Spectrometry Imaging and In Situ Top-Down Identification of Intact Proteins Directly from Tissue | Journal of the American Society for Mass Spectrometry
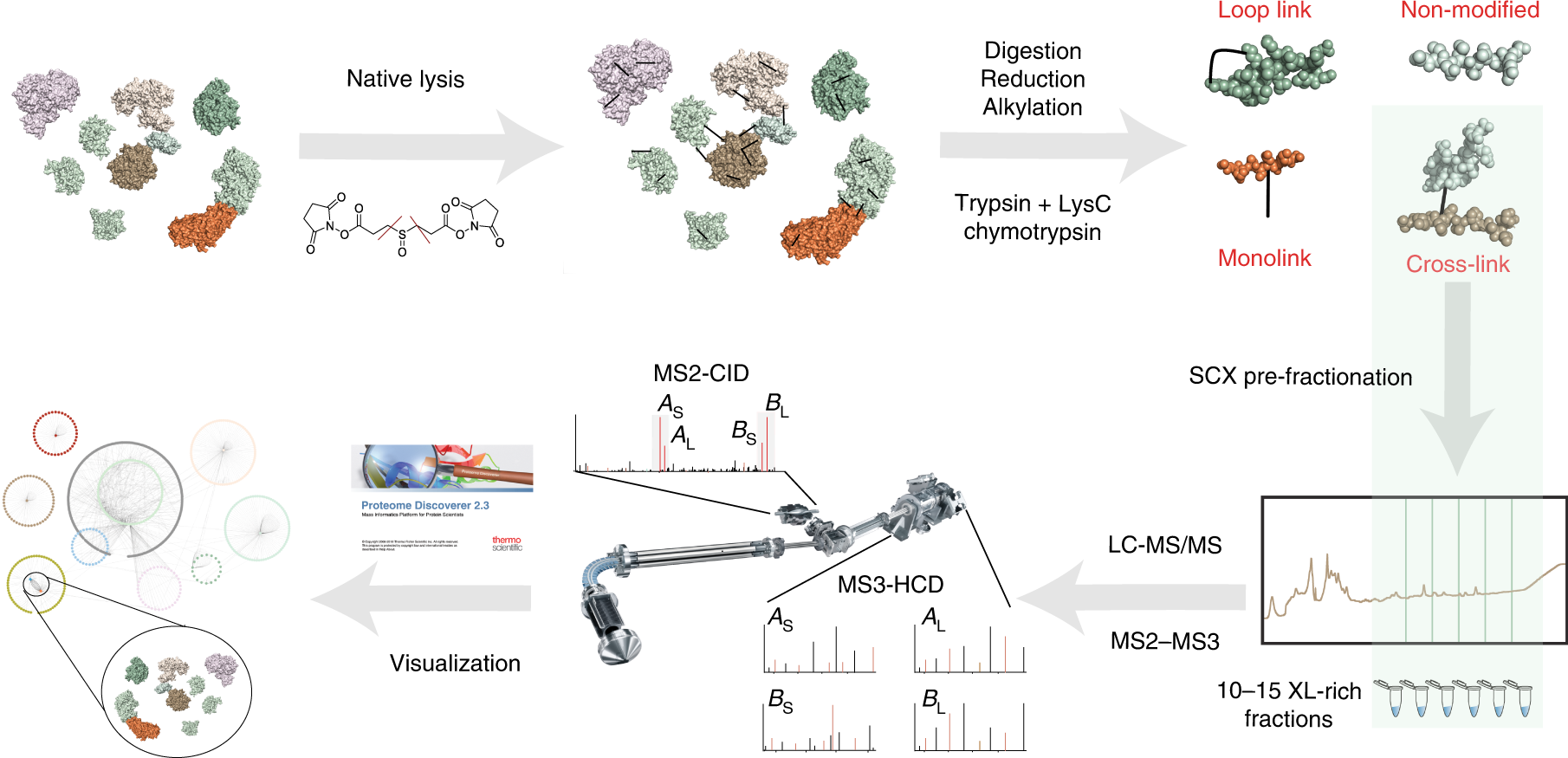
Efficient and robust proteome-wide approaches for cross-linking mass spectrometry | Nature Protocols

Characterization of an A3G-VifHIV-1-CRL5-CBFβ Structure Using a Cross-linking Mass Spectrometry Pipeline for Integrative Modeling of Host–Pathogen Complexes - ScienceDirect
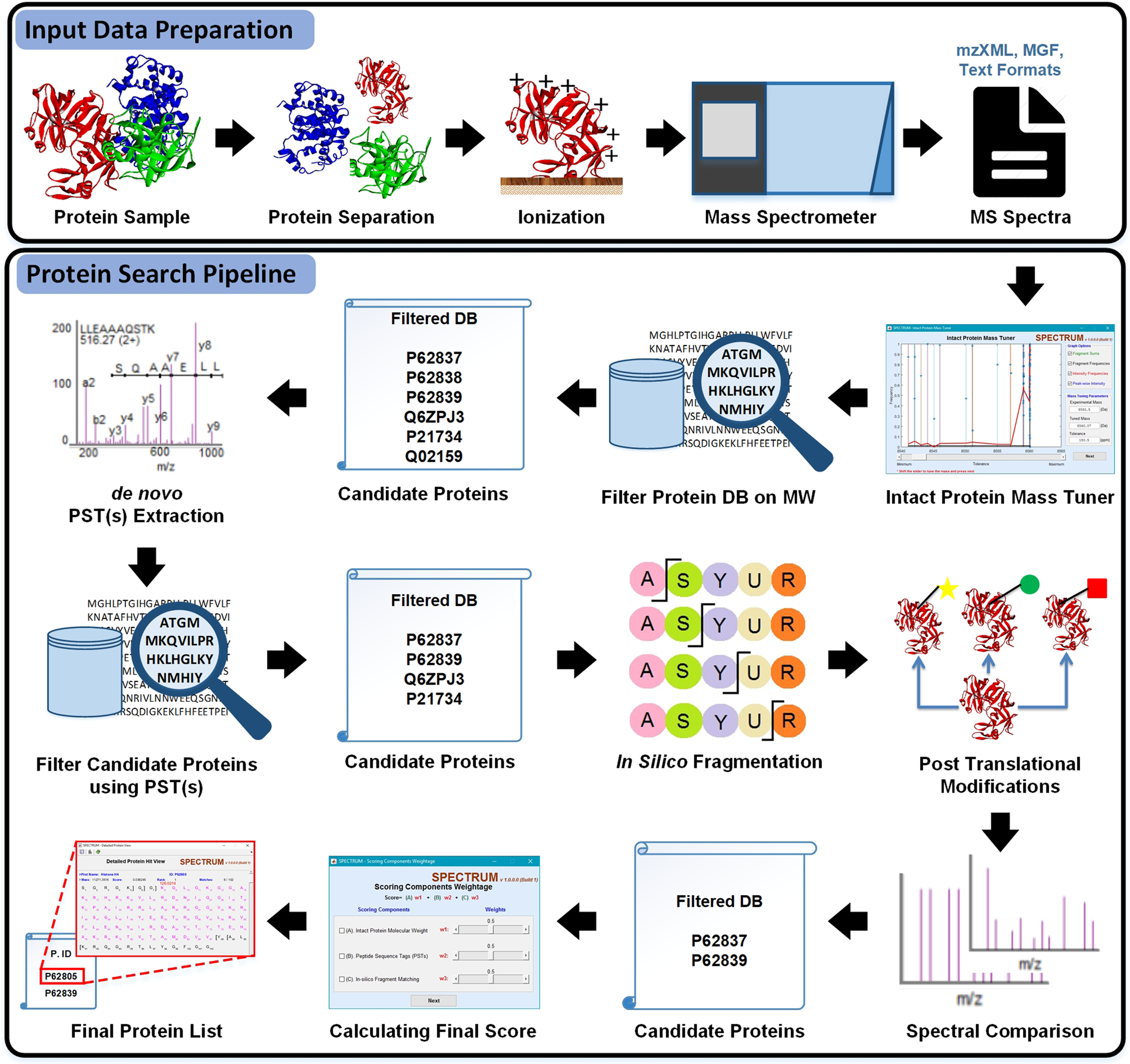
SPECTRUM – A MATLAB Toolbox for Proteoform Identification from Top-Down Proteomics Data | Scientific Reports

Discovery of Protein Modifications Using Differential Tandem Mass Spectrometry Proteomics | Journal of Proteome Research
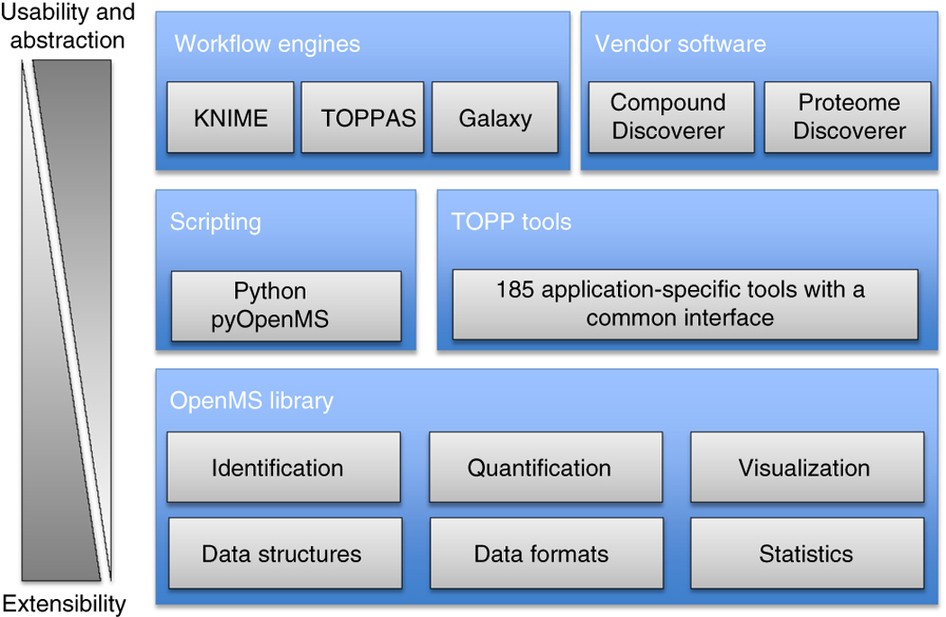
OpenMS: a flexible open-source software platform for mass spectrometry data analysis | Nature Methods

compareMS2 2.0: An Improved Software for Comparing Tandem Mass Spectrometry Datasets | Journal of Proteome Research

Determining Alternative Protein Isoform Expression Using RNA Sequencing and Mass Spectrometry - ScienceDirect
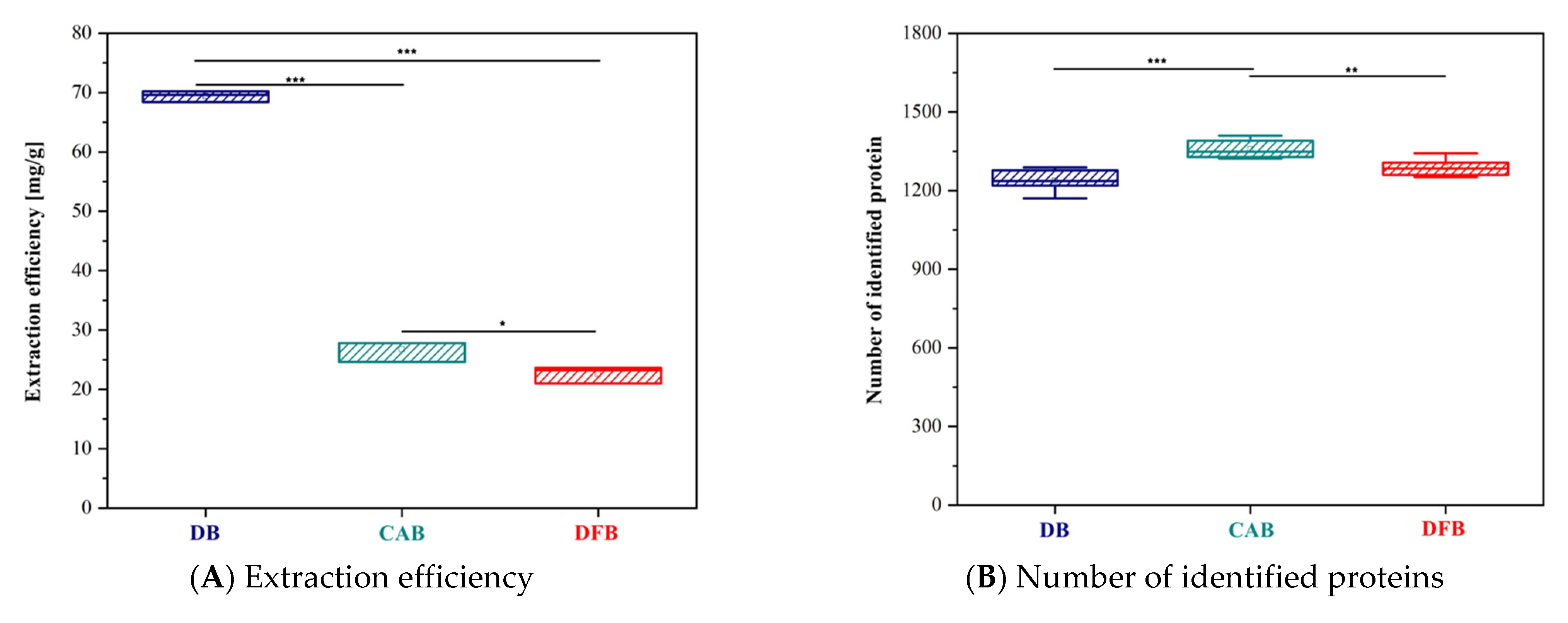
Biomedicines | Free Full-Text | Study on Tissue Homogenization Buffer Composition for Brain Mass Spectrometry-Based Proteomics